Local researchers said they have built a three-dimensional (3D) cancer genome map, which they said was the world’s largest.
On Monday, the research team of Professor Jung In-kyung of biological sciences at the Korea Advanced Institute of Science and Technology (KAIST) and Professor Lee Byung-wook at the Korean Bioinformation Center of Korea Research Institute of Bioscience and Biotechnology (KRIBB) said they have successfully created the world’s largest 3D cancer genome database.
The results will provide an important clue to understanding the pathogenesis of cancer and anticancer drug development, the research team said.
The researchers collected genome data of normal cell lines and tissues and cancer cell lines and tissues. Then, they constructed over 400 kinds of 3D human genome maps. The research team proposed a new strategy that can interpret a large-scale structural variation that occurs in cancer cells frequently.
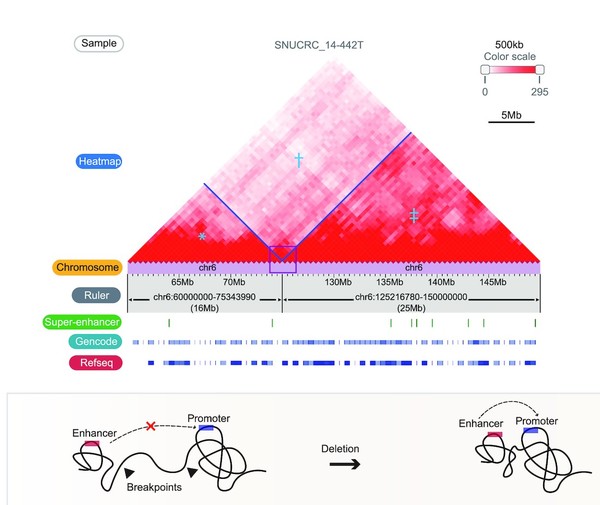
So far, researchers have continuously attempted to identify the cause of cancer by discovering mutations in the genome of cancer cells. More recently, unraveling a particular gene expression regulation mechanism in new cancer cells through large-scale structural variations has become an important research subject.
However, most structural variations exist in the non-transcribed areas where DNA does not produce proteins. So, identifying the mechanism through one-dimensional genomic sequence analysis had a limitation.
The research team designed the study based on the thought that it might be possible to investigate the gene expression regulation if they could uncover the “chromatin loop,” created and lost by large-scale structural variations, through a 3D genome structure decoding.
“This study showed that the function of structural variations that frequently occur in cancer could be accurately identified through 3D genome mapping,” Jung said.
“The results will be an opportunity to upgrade the technology to decode the cancer genome, which has not been fully identified.”
The National Research Foundation and Suh Kyung-bae Science Foundation supported the study.

